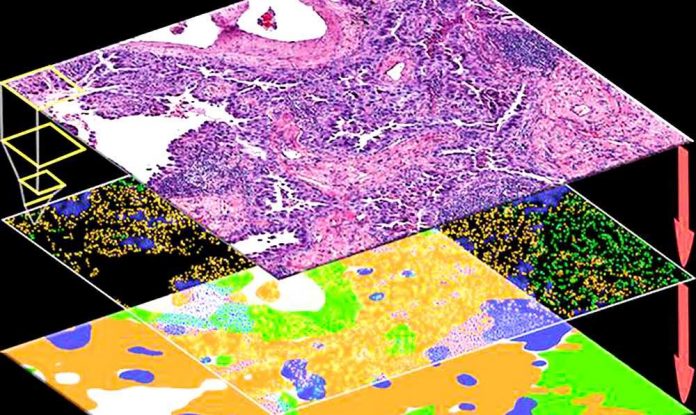
Researchers at the University of Chicago Medicine Comprehensive Cancer Center, working with colleagues in Europe, created a deep learning algorithm that can infer molecular alterations directly from routine histology images across multiple common tumor types.
The study, published July 27 in Nature Cancer, highlights the potential of artificial intelligence to help clinicians make personalized treatment plans for patients based on the information gained from how tissues appear under the microscope.
“We found that using artificial intelligence, we can quickly and accurately screen cancer patient biopsies for certain genetic alterations that may inform their treatment options and likelihood to respond to specific therapies,” said co-corresponding author Alexander Pearson, MD, PhD, assistant professor of medicine at UChicago Medicine.
“We are able to detect these genetic alterations almost instantly from a single slide, instead of requiring additional testing post-biopsy,” said Pearson. “If this model was validated and deployed at scale, it could dramatically improve the speed of molecular diagnosis across many cancers.”
Pearson and colleagues noted that although comprehensive molecular and genetic tests are difficult to implement at scale, tissue sections stained and mounted on a slide are commonplace and easy to study. And because molecular alterations in cancer can cause observable changes in tumor cells and their microenvironment, the researchers hypothesized that these structural changes would be visible on images of tissue slices captured under the microscope. In other words, genotype, the genetic make-up of the tumor cells, including gene mutations in key oncogenic pathways, influences the visible traits of those cells, known as their phenotype.
To test this, they set out to systematically investigate the presence of genotype-phenotype links for a wide range of clinically relevant molecular features across all major solid tumor types. Specifically, they asked which molecular features leave a strong enough footprint in histomorphology that they can be determined from histology images alone with deep learning.
To this end, the research team developed, optimized and externally validated a one-stop-shop workflow to train and evaluate deep learning networks to detect any sequence variants in these target genes. Leveraging The Cancer Genome Atlas, they applied this approach to hundreds of molecular alterations in tissue slides of more than 5,000 patients across 14 major tumor types.
They found that in 13 out of 14 tested tumor types, the mutation of one or more such genes could be inferred from histology images alone. In particular, in major cancer types such as lung, colorectal, breast and gastric cancers, alterations of several genes of particular clinical interest were detectable.
Examples include mutations in TP53, which could be significantly detected in all four of these cancer types, as well as mutations of BRAF in colorectal cancer. Among all tested tumor types, gastric cancer and colorectal cancer had the highest absolute number of detectable mutations.
In addition, they found that higher-level gene expression clusters or signatures can be inferred from histological images. These findings that molecular signatures of tumors reflect biologically distinct groups and are correlated with clinical outcome could open up new options for personalized cancer treatment.
Another research group has independently validated these results with a similar AI algorithm applied to images from common cancer types. Their study was published in the same issue of Nature Cancer.