An international research team has for the first time obtained the structure of the light-sensitive sodium-pumping KR2 protein in its active state. The discovery provides a description of the mechanism behind the light-driven sodium ion transfer across the cell membrane. The paper came out in Nature Communications.
KR2 is a member of a very large family of microbial rhodopsins — light-sensitive proteins present in the cell membrane of archaea, bacteria, viruses, and eukaryotes. These proteins have a wide range of functions, including light-driven transport of ions across the membrane. Such ion channels and pumps are the primary tools of optogenetics, a booming field in biomedicine with a focus on controlling cells in the body by illuminating them with light.
Optogenetics came to prominence due to its contributions to minimally invasive techniques for brain research and neurodegenerative disorder treatments addressing Alzheimer’s, Parkinson’s, and other diseases. Beyond that, optogenetics enables reversing vision and hearing loss and restoring muscle activity.
Despite its many successes, further development of optogenetics is complicated by the limited number of available proteins suitable for cell activation and inhibition. For example, the most widely used optogenetic tool, channelrhodopsin 2, whose structure was originally reported in Science by MIPT researchers and graduates, can transport both sodium, potassium, and calcium ions, as well as protons. The protein’s low selectivity leads to undesirable side effects on cells. As a result, optimizing the protocols for using optogenetic tools is currently costly and time-intensive.
The search for new, more selective proteins is a priority for optogenetics. One of the candidates, the KR2 rhodopsin discovered in 2013, is a unique tool that selectively transports only the sodium ions across the membrane under physiological conditions. Understanding how KR2 works is crucial for optimizing the functional characteristics of that protein and using it as the basis for new optogenetic tools.
MIPT biophysicists published the first structures of KR2 in its various forms in 2015 and 2019. Among other things, they showed that the protein organizes into pentamers in the membrane, and that such behavior is vital to its functioning.
However, all the models described so far have looked at the protein in its inactive, or ground state. Yet it is only in the active state — after illumination — that the protein actually transports sodium. To understand how the KR2 pump works, the researchers have now obtained and described its high-resolution structure in the active state.
“We began by using the traditional approach, activating KR2 in pregrown protein crystals by illuminating them with a laser and getting a snapshot of the active state by rapidly freezing the crystals at 100 kelvins,” said the study’s first author, MIPT doctoral student Kirill Kovalev. “We got lucky, because such manipulations may well destroy the crystals. To avoid this, we had to fine-tune the laser wavelength and power and find the optimal exposure time.”
Producing the large number of high-quality KR2 rhodopsin crystals necessary for the experiments has been made possible by the unique equipment of the MIPT Research Center for Molecular Mechanisms of Aging and Age-Related Diseases.
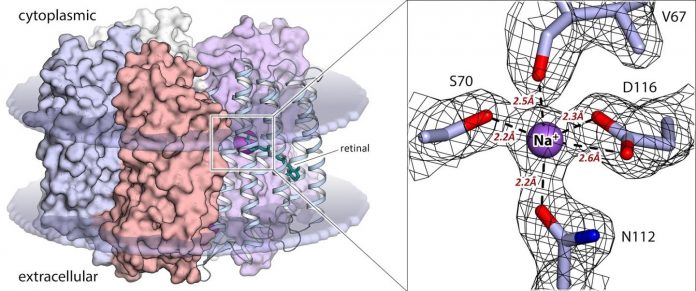
The most significant finding of the study is identifying the amino acid residues of the protein that bind the sodium ion inside the KR2 molecule. They are the factor that determines the rhodopsin selectivity toward a particular type of ions. In addition to that, a high-resolution structure for the protein’s active state at 2.1 angstroms — 21 hundred-billionths of a meter — has revealed the precise configuration of the sodium ion binding site at the protein’s active center. For the first time, the team showed that the binding site of KR2 has become optimized for sodium ions in the course of rhodopsin evolution. This means that the active state structure obtained in the study is best-suited for the rational design of next-generation KR2-based optogenetic tools.
“In the course of our work, we also obtained the active-state KR2 structure at room temperature,” Kovalev added. “To achieve this, we had to update the well-known protocols for collecting crystallographic data. Besides, we employed a synchrotron radiation source to leverage the serial crystallography techniques, which are growing popular right now.”
The room temperature KR2 structure confirmed that the protein model produced from a low-temperature snapshot is correct. This provided a direct demonstration that cryogenic freezing did not affect the rhodopsin’s internal structure.
The structures reported in the paper have allowed the scientists to provide a first-ever description of active light-driven sodium ion transport across the cell membrane. Specifically, the study shows that sodium transport most likely involves a hybrid mechanism comprised by relay proton transport and passive ion diffusion through polar cavities in the protein. The mechanism proposed by the researchers has been confirmed via functional studies of mutated KR2 forms and molecular dynamics simulations of sodium ion release from the protein.
“Ion transport across the cell membrane is a fundamental biological process. That said, sodium ion transport should be enabled by a mechanism distinct from that involved in proton transport,” explains Valentin Gordeliy, the director for research at the Grenoble institute for Structural Biology and the scientific coordinator of the MIPT Research Center for Molecular Mechanisms of Aging and Age-Related Diseases. “For the first time, we see how a sodium ion is bound inside the rhodopsin molecule and understand the mechanism for ion release into the intercellular space.”
The biophysicists are convinced that their findings not only reveal the fundamental principles underlying ion transport across the membrane but will be of use to optogenetics. MIPT is continuing the development of optimized KR2 protein forms to expand the toolkit for brain research and neurodegenerative disease therapies.