Researchers from the Moscow Institute of Physics and Technology, working with Spanish, French, and German colleagues, have determined and analyzed the high-resolution structure of a protein from the recently discovered heliorhodopsin family. Microbial rhodopsins play a key role in optogenetics — a technique that uses light to control nerve and muscle cells in living tissue. The findings were published in PNAS.
Optogenetics, a technique developed in 2005, involves the introduction of special photosensitive proteins, rhodopsins, into the membranes of neurons. It presents new possibilities for treating Parkinson’s disease, depression, anxiety, and epilepsy.
Rhodopsins: Key tool for optogenetics
What made optogenetics possible was the discovery of a protein subfamily known as channelrhodopsins. Rhodopsins are activated by light, which they capture using a compound called retinal. That property makes rhodopsins crucial for animal vision.
In the 20th century, only archaeal rhodopsins found in the genomes of halobacteria were known. However, genomic and metagenomic studies have since led to the discovery of more than 10,000 rhodopsin genes across all three domains of life — bacteria, archaea, and eukaryotes — as well as in giant viruses. Among other things, these proteins are responsible for most of the solar energy captured in the ocean.
The diversity, biological significance, and possibilities for the applied use of rhodopsins prompt research into their structure. This in turn provides insights into their mechanisms of action and functions. Scientists from the Research Center for Molecular Mechanisms of Aging and Age-Related Diseases at MIPT make regular contributions to the field. Last year, for instance, they determined the structure of the KR2 protein as well as that of a rhodopsin from a giant virus.
‘Inverted’ protein structure examined
Despite the many differences in structure and function among rhodopsins, their orientation in the membrane is usually the same, with the so-called N-terminus positioned on the outside of the cell. The N-terminus refers to the end of a protein carrying an amino group, the other end being the C-terminus, carrying a carboxyl group.
However, in the recently discovered family of heliorhodopsins, it is the other way around: The N-terminus faces the cytoplasm, that is, the inside of the cell.
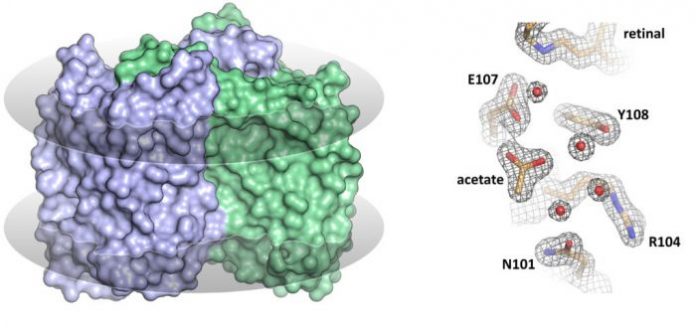
The MIPT researchers are among the first to have solved the structure of a heliorhodopsin — namely, the 48C12 protein (fig. 1) — showing the key differences from other known rhodopsins. In the recent paper, the team also suggests the possible functions of heliorhodopsins.
Kirill Kovalev, one of the first authors of the study and MIPT doctoral student, said: “Heliorhodopsins are unusual proteins. The high-resolution structures we obtained demonstrate both their unique overall architecture and the details of their internal configuration and the interactions between key amino acid residues.”
In their study, the researchers analyzed the structure of the 48C12 protein in two states and compared it with the structures of other microbial rhodopsins. The team demonstrated that there is a large cavity inside the protein’s cytoplasmic part, which accommodates a cluster of water molecules. In one of the obtained protein states, an acetate molecule was detected in the cavity. This means that the cavity might act as the protein’s active site, where the binding of a substrate such as a nitrate or a carbonate occurs.
Though the precise function and biological role of heliorhodopsins remain unknown, the authors have put forward a hypothesis. “The unusual structure and properties of the protein leads us to suggest an enzymatic function for heliorhodopsins. Our work has also shown that protein groups with distinct functions may be identified within the family,” explained Valentin Gordeliy, the scientific coordinator of MIPT’s Research Center for Molecular Mechanisms of Aging and Age-Related Diseases.
The discovery of the heliorhodopsin 48C12 structure has thus allowed the biologists to demonstrate its fundamental difference from other microbial rhodopsins, opening up new possibilities for further research into heliorhodopsins.